|
DIVULGAÇÃO
What is “DNA origami” and how does it help develop vaccines ? DNA origami is a technique by which DNA can be folded into 2D and 3D structures at the nanoscale. In 2016, MIT researchers used this technique to develop an algorithm that could automatically design and build arbitrary 3D virus-like shapes, allowing for attaching a variety of molecules such as viral antigens at specific locations on the DNA. The researchers could design HIV-like particles that provoked a strong immune response from lab-grown human immune cells, and recently, they swapped out the HIV antigens for a protein found on the surface of the SARS-CoV-2 virus to develop a viable vaccine candidate for COVID-19. 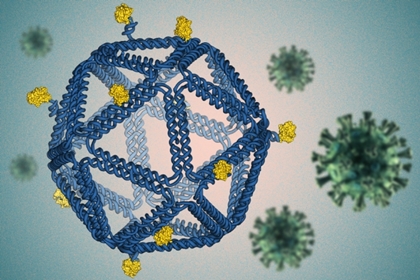 By folding DNA into a virus-like structure, MIT researchers have designed HIV-like particles that provoke a strong immune response from human immune cells grown in a lab dish. Image: Courtesy of the researchers, edited by MIT News
“The rough design rules that are starting to come out of this work should be generically applicable across disease antigens and diseases,” says Darrell Irvine, who is the Underwood-Prescott Professor with appointments in the departments of Biological Engineering and Materials Science and Engineering; an associate director of MIT’s Koch Institute for Integrative Cancer Research; and a member of the Ragon Institute of MGH, MIT, and Harvard. Irvine and Mark Bathe, an MIT professor of biological engineering and an associate member of the Broad Institute of MIT and Harvard, are the senior authors of the study, which appears today in Nature Nanotechnology. The paper’s lead authors are former MIT postdocs Rémi Veneziano and Tyson Moyer. In 2016, Bathe’s lab developed an algorithm that can automatically design and build arbitrary three-dimensional virus-like shapes using DNA origami. This method offers precise control over the structure of synthetic DNA, allowing researchers to attach a variety of molecules, such as viral antigens, at specific locations “The DNA structure is like a pegboard where the antigens can be attached at any position,” Bathe says. “These virus-like particles have now enabled us to reveal fundamental molecular principles of immune cell recognition for the first time.” Natural viruses are nanoparticles with antigens arrayed on the particle surface, and it is thought that the immune system (especially B cells) has evolved to efficiently recognize such particulate antigens. Vaccines are now being developed to mimic natural viral structures, and such nanoparticle vaccines are believed to be very effective at producing a B cell immune response because they are the right size to be carried to the lymphatic vessels, which send them directly to B cells waiting in the lymph nodes. The particles are also the right size to interact with B cells and can present a dense array of viral particles. However, determining the right particle size, spacing between antigens, and number of antigens per particle to optimally stimulate B cells (which bind to target antigens through their B cell receptors) has been a challenge. Bathe and Irvine set out to use these DNA scaffolds to mimic such viral and vaccine particle structures, in hopes of discovering the best particle designs for B cell activation. “There is a lot of interest in the use of virus-like particle structures, where you take a vaccine antigen and array it on the surface of a particle, to drive optimal B-cell responses,” Irvine says. “However, the rules for how to design that display are really not well-understood.” Other researchers have tried to create subunit vaccines using other kinds of synthetic particles, such as polymers, liposomes, or self-assembling proteins, but with those materials, it is not possible to control the placement of viral proteins as precisely as with DNA origami. For this study, the researchers designed icosahedral particles with a similar size and shape as a typical virus. They attached an engineered HIV antigen related to the gp120 protein to the scaffold at a variety of distances and densities. To their surprise, they found that the vaccines that produced the strongest response B cell responses were not necessarily those that packed the antigens as closely as possible on the scaffold surface. “It is often assumed that the higher the antigen density, the better, with the idea that bringing B cell receptors as close together as possible is what drives signaling. However, the experimental result, which was very clear, was that actually the closest possible spacing we could make was not the best. And, and as you widen the distance between two antigens, signaling increased,” Irvine says. The findings from this study have the potential to guide HIV vaccine development, as the HIV antigen used in these studies is currently being tested in a clinical trial in humans, using a protein nanoparticle scaffold. Based on their data, the MIT researchers worked with Jayajit Das, a professor of immunology and microbiology at Ohio State University, to develop a model to explain why greater distances between antigens produce better results. When antigens bind to receptors on the surface of B cells, the activated receptors crosslink with each other inside the cell, enhancing their response. However, the model suggests that if the antigens are too close together, this response is diminished. “Our platform technology allows you to easily swap out different subunit antigens and peptides from different types of viruses to test whether they may potentially be functional as vaccines,” Bathe says. Because this approach allows for antigens from different viruses to be carried on the same DNA scaffold, it could be possible to design variants that target multiple types of coronaviruses, including past and potentially future variants that may emerge, the researchers say. Bathe was recently awarded a grant from the Fast Grants Covid-19 fund to develop their SARS-CoV-2 vaccine. The HIV research presented in the Nature Nanotechnology paper was funded by the Human Frontier Science Program, the U.S. Office of Naval Research, the U.S. Army Research Office through MIT’s Institute for Soldier Nanotechnologies, the Ragon Institute, and the U.S. National Institutes of Health. Read the original article on MIT News. Accessed: July 16, 2020.
|
||||||||||||||||||||||||